Example queries
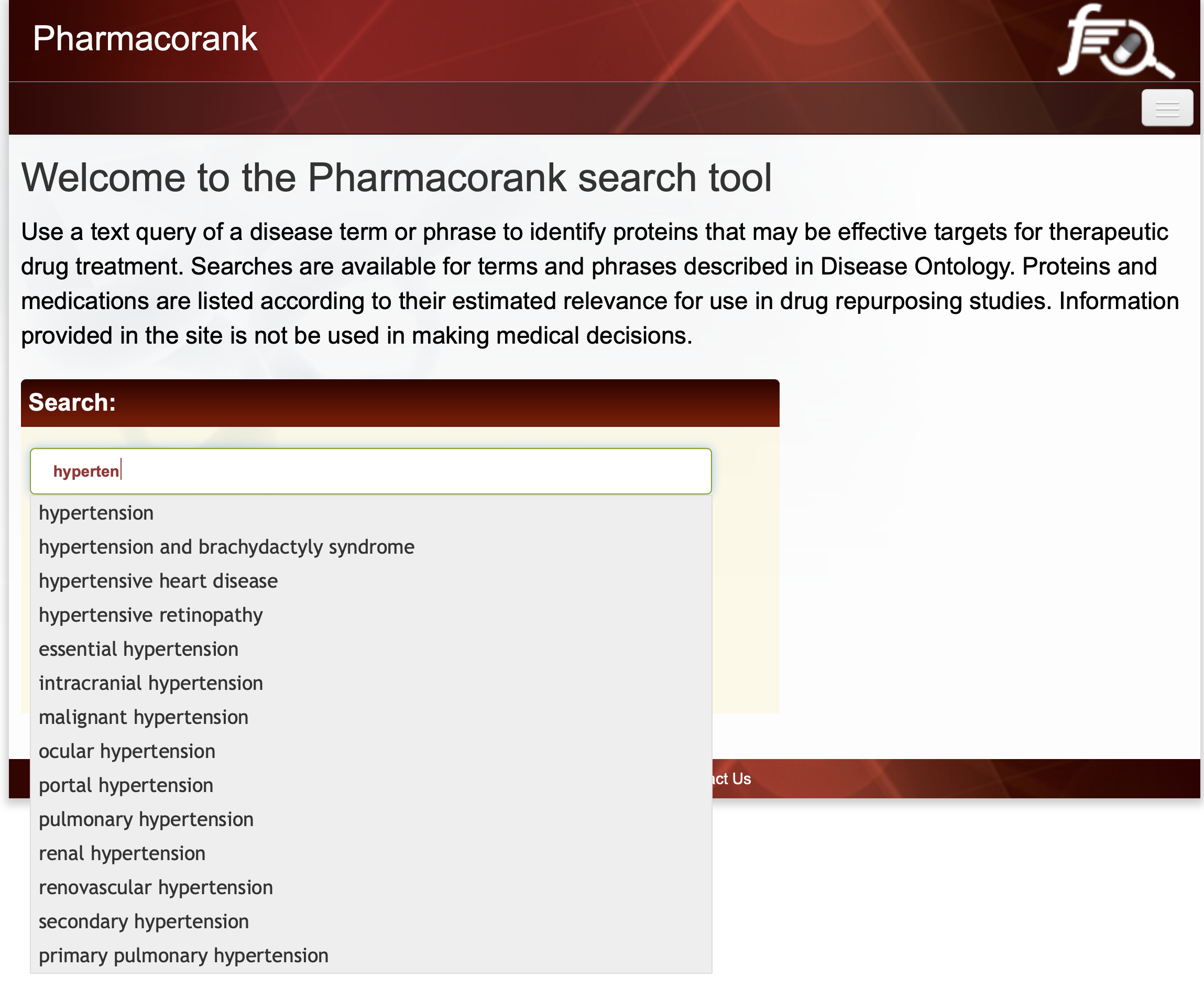
To initiate an interactive query, begin typing a disease name in the query input box. The corresponding Disease Ontology term will be identified with an autocomplete feature if the disease was found to have associated proteins. Here we review the results when the query term is hypertension. Shown in Figure 1 is the autocomplete tool identifying possible search terms based on the user’s input.
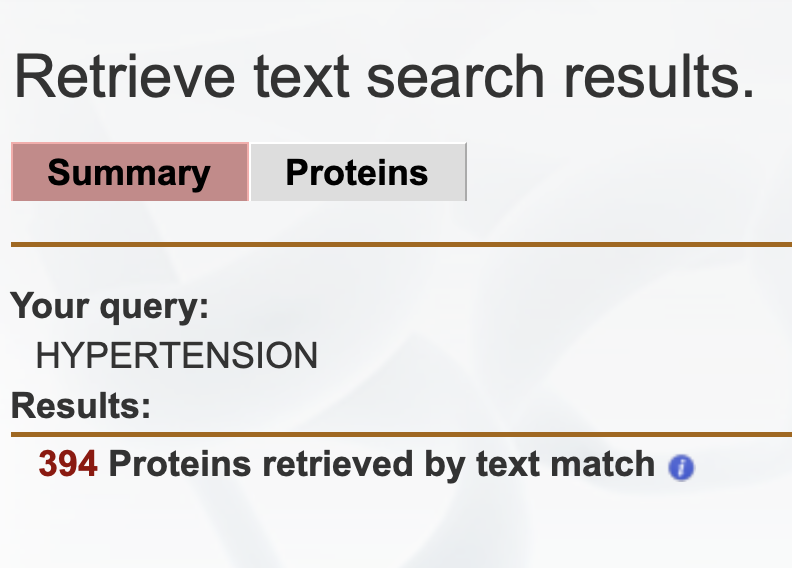
After clicking on the search button, a summary of the prioritized list of target proteins retrieved is displayed, as shown in Figure 2. The total number of proteins retrieved by the text matching algorithm is displayed. One can click on the red number that indicates the total number of proteins retrieved and next be taken to a prioritized list of target proteins.
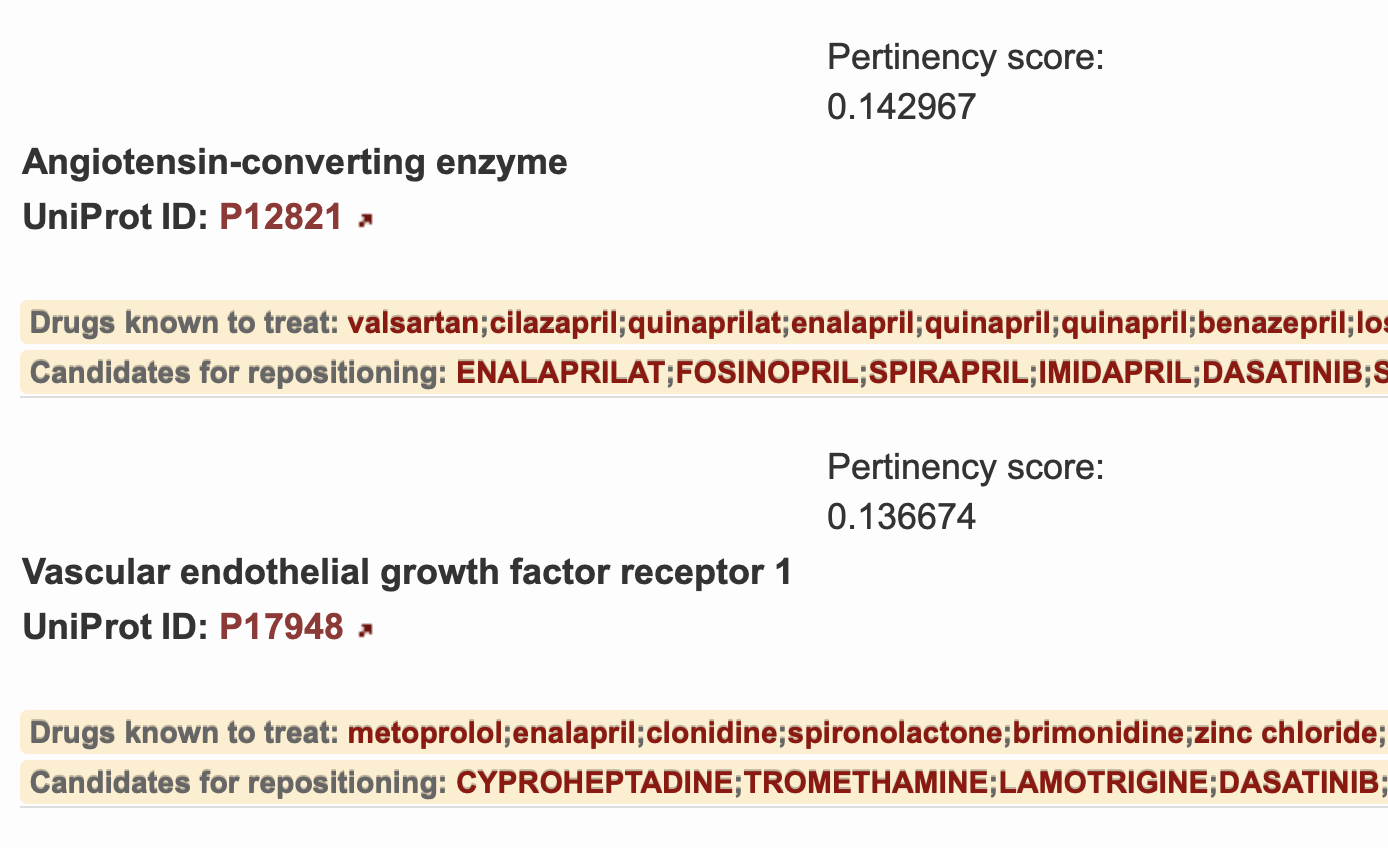
A prioritized list of proteins is shown in Figure 3. On the left, the accepted protein name is displayed and it's respective UniProt ID can be found immediately below it. The UniProt ID is linked to it's corresponding information page within the UniProtKB database. The calculated pertinency score is shown to the right of each protein target which estimates the probability that the protein target is relevant in the disease mechanism and the associated medications may be pertinent in the treatment of the selected disease. A label is provided for each medication as to whether it is known to treat or not known to treat the selected disease. Drugs not currently known to treat the disease are described as repositioning candidates.